Visual Interpretation of Principal Coordinates (of) Neighbor Matrices (PCNM)
Feb 21, 2010 metroadminPrincipal Coordinates (of) Neighbor Matrices (PCNM) is an interesting algorithm, developed by P. Borcard and P. Legendre at the University of Montreal, for the multi-scale analysis of spatial structure. This algorithm is typically applied to a distance matrix, computed from the coordinates where some environmental data were collected. The resulting "PCNM vectors" are commonly used to describe variable degrees of possible spatial structure and its contribution to variability in other measured parameters (soil properties, species distribution, etc.)-- essentially a spectral decomposition spatial connectivity. This algorithm has been recently updated by and released as part of the PCNM package for R. Several other implementations of the algorithm exist, however this seems to be the most up-to-date.
Related Presentations and Papers on PCNM
- http://biol09.biol.umontreal.ca/ESA_SS/Borcard_&_PL_talk.pdf
- Borcard, D. and Legendre, P. 2002. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153: 51-68.
- Borcard, D., P. Legendre, Avois-Jacquet, C. & Tuomisto, H. 2004. Dissecting the spatial structures of ecologial data at all scales. Ecology 85(7): 1826-1832.
I was interested in using PCNM vectors for soils-related studies, however I encountered some in difficulty interpreting what they really meant when applied to irregularly-spaced site locations. As a demonstration, I generated several (25 to be exact) PCNM vectors from a regular grid of points. Using an example from the PCNM manual page, I have plotted the values of the PCNM vectors at the grid nodes (below). The interpretation of the PCNM vectors derived from a 2D, regular grid is fairly simple: lower order vectors represent regional-scale groupings, higher order vectors represent more local-scale groupings. One thing to keep in mind is that these vectors give us a multi-scale metric for grouping sites, and are not computed by any properties that may have been measured at the sites. The size of the symbols are proportional to the PCNM vectors and the color represents the sign.
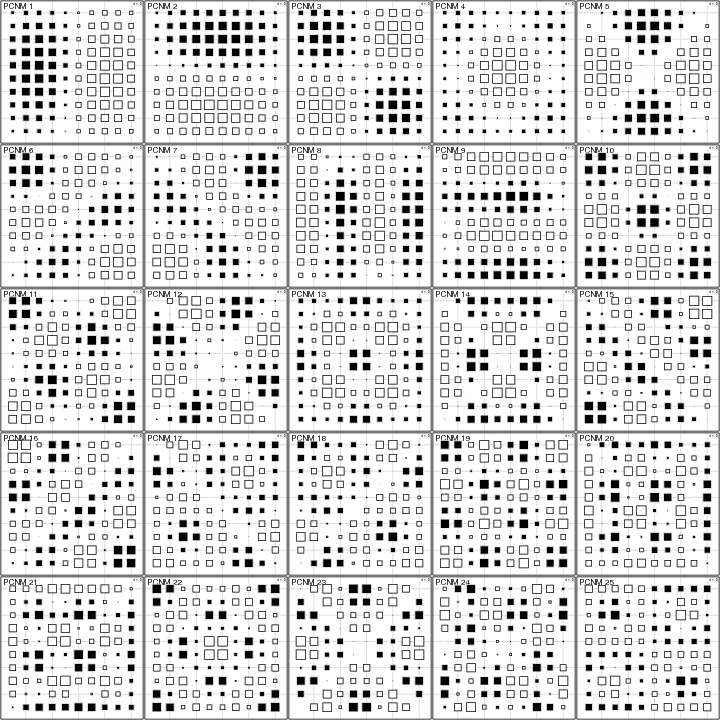
Figure: PCNM - Regular Grid
Soil survey operations are rarely conducted on a regular grid, so I re-computed PCNM vectors from the same simulated grid, but after randomly perturbing each site. The resulting map of PCNM vectors is presented below. The patterns are a little complex, but quickly decipherable with guidance from the PCNM vectors derived from a regular grid. Neat

Figure: PCNM - Randomly Perturbed Regular Grid
R code used to make figures
library(ade4)
library(PCNM)
# fake data
g <- expand.grid(x=1:10, y=1:10)
x.coords <- data.frame(x=g$x, y=g$y)
# PCNM
x.sub.dist <- dist(x.coords[,c('x','y')])
x.sub.pcnm <- PCNM(x.sub.dist, dbMEM=TRUE)
# plot first 25 PCNM vectors
pdf(file='PCNM-grid-example.pdf', width=10, height=10)
par(mfrow=c(5,5))
for(i in 1:25)
s.value(x.coords[,c('x','y')], x.sub.pcnm$vectors[,i], clegend=0, sub=paste("PCNM", i), csub=1.5, addaxes=FALSE, origin=c(1,1))
dev.off()
# jitter the same input and try again
x.coords <- data.frame(x=jitter(g$x, factor=2), y=jitter(g$y, factor=2))
x.sub.dist <- dist(x.coords[,c('x','y')])
x.sub.pcnm <- PCNM(x.sub.dist, dbMEM=TRUE)
# plot first 25 PCNM vectors
pdf(file='PCNM-jittered_grid-example.pdf', width=10, height=10)
par(mfrow=c(5,5))
for(i in 1:25)
s.value(x.coords[,c('x','y')], x.sub.pcnm$vectors[,i], clegend=0, sub=paste("PCNM", i), csub=1.5, addaxes=FALSE, origin=c(1,1))
dev.off()
Links:
Target Practice and Spatial Point Process Models
Working with Spatial Data
Visualizing Random Fields and Select Components of Spatial Autocorrelation
Software
- General Purpose Programming with Scripting Languages
- LaTeX Tips and Tricks
- PostGIS: Spatially enabled Relational Database Sytem
- PROJ: forward and reverse geographic projections
- GDAL and OGR: geodata conversion and re-projection tools
- R: advanced statistical package
- Access Data Stored in a Postgresql Database
- Additive Time Series Decomposition in R: Soil Moisture and Temperature Data
- Aggregating SSURGO Data in R
- Cluster Analysis 1: finding groups in a randomly generated 2-dimensional dataset
- Color Functions
- Comparison of Slope and Intercept Terms for Multi-Level Model
- Comparison of Slope and Intercept Terms for Multi-Level Model II: Using Contrasts
- Creating a Custom Panel Function (R - Lattice Graphics)
- Customized Scatterplot Ideas
- Estimating Missing Data with aregImpute() {R}
- Exploration of Multivariate Data
- Interactive 3D plots with the rgl package
- Making Soil Property vs. Depth Plots
- Numerical Integration/Differentiation in R: FTIR Spectra
- Plotting XRD (X-Ray Diffraction) Data
- Using lm() and predict() to apply a standard curve to Analytical Data
- Working with Spatial Data
- Customizing Maps in R: spplot() and latticeExtra functions
- Converting Alpha-Shapes into SP Objects
- Some Ideas on Interpolation of Categorical Data
- Visual Interpretation of Principal Coordinates (of) Neighbor Matrices (PCNM)
- Visualizing Random Fields and Select Components of Spatial Autocorrelation
- Generation of Sample Site Locations [sp package for R]
- Target Practice and Spatial Point Process Models
- Ordinary Kriging Example: GRASS-R Bindings
- Point-process modelling with the sp and spatstat packages
- Simple Map Creation
- Comparison of PSA Results: Pipette vs. Laser Granulometer
- GRASS GIS: raster, vector, and imagery analysis
- Generic Mapping Tools: high quality map production

