Aggregating SSURGO Data in R
Sep 10, 2009 metroadminPremise
SSURGO is a digital, high-resolution (1:24,000), soil survey database produced by the USDA-NRCS. It is one of the largest and most complete spatial databases in the world; and is available for nearly the entire USA at no cost. These data are distributed as a combination of geographic and text data, representing soil map units and their associated properties. Unfortunately the text files do not come with column headers, so a template is required to make sense of the data. Alternatively, one can use an MS Access templateto attach column names, generate reports, and other such tasks. CSV file can be exported from the MS Access database for further use. A follow-up post with text file headers, and complete PostgreSQL database schema will contain details on implementing a SSURGO database without using MS Access.
If you happen to have some of the SSURGO tabular data that includes column names, the following R code may be of general interest for resolving the 1:many:many hierarchy of relationships required to make a thematic map.
This is the format we want the data to be in
mukey clay silt sand water_storage 458581 20.93750 20.832237 20.861842 14.460000 458584 43.11513 30.184868 26.700000 23.490000 458593 50.00000 27.900000 22.100000 22.800000 458595 34.04605 14.867763 11.776974 18.900000
So we can make a map like this
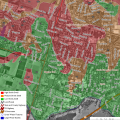
Loading Data Into R
# need this for ddply()
library(plyr)
# load horizon and component data
chorizon <- read.csv('chorizon_table.csv')
# only keep some of the columns from the component table
component <- read.csv('component_table.csv')[,c('mukey','cokey','comppct_r')]
Function Definitions
# custom function for calculating a weighted mean
# values passed in should be vectors of equal length
wt_mean <- function(property, weights)
{
# compute thickness weighted mean, but only when we have enough data
# in that case return NA
# save indices of data that is there
property.that.is.na <- which( is.na(property) )
property.that.is.not.na <- which( !is.na(property) )
if( length(property) - length(property.that.is.na) >= 1)
prop.aggregated <- sum(weights[property.that.is.not.na] * property[property.that.is.not.na], na.rm=TRUE) / sum(weights[property.that.is.not.na], na.rm=TRUE)
else
prop.aggregated <- NA
return(prop.aggregated)
}
profile_total <- function(property, thickness)
{
# compute profile total
# in that case return NA
# save indices of data that is there
property.that.is.na <- which( is.na(property) )
property.that.is.not.na <- which( !is.na(property) )
if( length(property) - length(property.that.is.na) >= 1)
prop.aggregated <- sum(thickness[property.that.is.not.na] * property[property.that.is.not.na], na.rm=TRUE)
else
prop.aggregated <- NA
return(prop.aggregated)
}
# define a function to perfom hz-thickness weighted aggregtion
component_level_aggregation <- function(i)
{
# horizon thickness is our weighting vector
hz_thick <- i$hzdepb_r - i$hzdept_r
# compute wt.mean aggregate values
clay <- wt_mean(i$claytotal_r, hz_thick)
silt <- wt_mean(i$silttotal_r, hz_thick)
sand <- wt_mean(i$sandtotal_r, hz_thick)
# compute profile sum values
water_storage <- profile_total(i$awc_r, hz_thick)
# make a new dataframe out of the aggregate values
d <- data.frame(cokey=unique(i$cokey), clay=clay, silt=silt, sand=sand, water_storage=water_storage)
return(d)
}
mapunit_level_aggregation <- function(i)
{
# component percentage is our weighting vector
comppct <- i$comppct_r
# wt. mean by component percent
clay <- wt_mean(i$clay, comppct)
silt <- wt_mean(i$silt, comppct)
sand <- wt_mean(i$sand, comppct)
water_storage <- wt_mean(i$water_storage, comppct)
# make a new dataframe out of the aggregate values
d <- data.frame(mukey=unique(i$mukey), clay=clay, silt=silt, sand=sand, water_storage=water_storage)
return(d)
}
Performing the Aggregation
# aggregate horizon data to the component level chorizon.agg <- ddply(chorizon, .(cokey), .fun=component_level_aggregation, .progress='text') # join up the aggregate chorizon data to the component table comp.merged <- merge(component, chorizon.agg, by='cokey') # aggregate component data to the map unit level component.agg <- ddply(comp.merged, .(mukey), .fun=mapunit_level_aggregation, .progress='text') # save data back to CSV write.csv(component.agg, file='something.csv', row.names=FALSE)
Attachment:
Links:
Additive Time Series Decomposition in R: Soil Moisture and Temperature Data
R: advanced statistical package
Cluster Analysis 1: finding groups in a randomly generated 2-dimensional dataset
Software
- General Purpose Programming with Scripting Languages
- LaTeX Tips and Tricks
- PostGIS: Spatially enabled Relational Database Sytem
- PROJ: forward and reverse geographic projections
- GDAL and OGR: geodata conversion and re-projection tools
- R: advanced statistical package
- Access Data Stored in a Postgresql Database
- Additive Time Series Decomposition in R: Soil Moisture and Temperature Data
- Aggregating SSURGO Data in R
- Cluster Analysis 1: finding groups in a randomly generated 2-dimensional dataset
- Color Functions
- Comparison of Slope and Intercept Terms for Multi-Level Model
- Comparison of Slope and Intercept Terms for Multi-Level Model II: Using Contrasts
- Creating a Custom Panel Function (R - Lattice Graphics)
- Customized Scatterplot Ideas
- Estimating Missing Data with aregImpute() {R}
- Exploration of Multivariate Data
- Interactive 3D plots with the rgl package
- Making Soil Property vs. Depth Plots
- Numerical Integration/Differentiation in R: FTIR Spectra
- Plotting XRD (X-Ray Diffraction) Data
- Using lm() and predict() to apply a standard curve to Analytical Data
- Working with Spatial Data
- Comparison of PSA Results: Pipette vs. Laser Granulometer
- GRASS GIS: raster, vector, and imagery analysis
- Generic Mapping Tools: high quality map production

